Research Highlight
APEX2 Enhances Proximity Labeling and Electron Microscopy Imaging to Resolve Dispute about Regulation of the Inner Membrane Channel
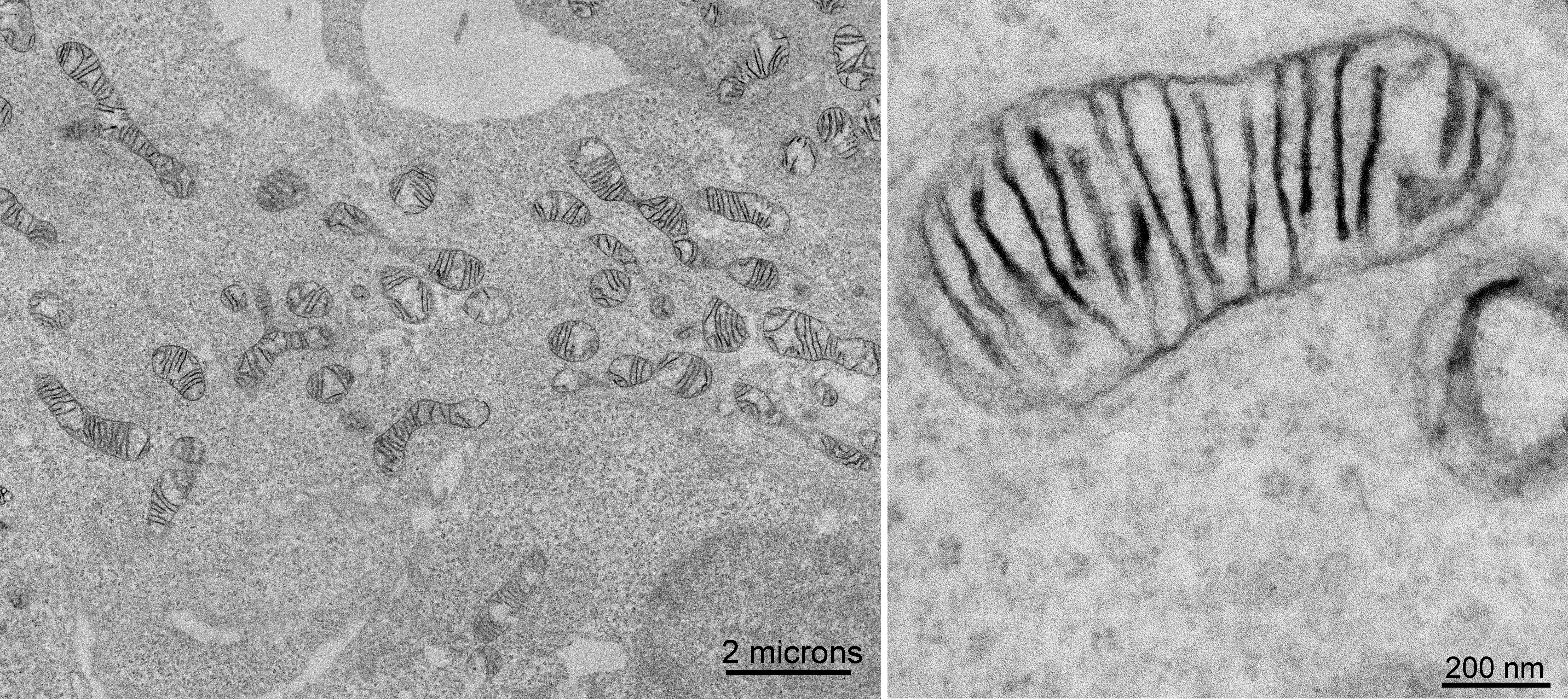 Figure Caption: (Left hand image) Electron micrograph of cultured human embryonic kidney cells stably expressing MICU1-APEX2 and reacted with diaminobenzidine to form a reaction product. (Right hand image) Higher magnification of a representative mitochondrion showed that staining is associated with the inner mitochondrial membranes rather than filling the intermembrane space.
Figure Caption: (Left hand image) Electron micrograph of cultured human embryonic kidney cells stably expressing MICU1-APEX2 and reacted with diaminobenzidine to form a reaction product. (Right hand image) Higher magnification of a representative mitochondrion showed that staining is associated with the inner mitochondrial membranes rather than filling the intermembrane space.
January 2015 La Jolla -- A research team from MIT, Massachusetts General Hospital, and UCSD in late November 2014 published a study in Nature Methods describing an electron microscopy tag, APEX2, which they developed for live-cell proteomics and genetic probe-based labeling for light and electron microscopy. Use of this tag enabled them to resolve a conflict between competing mechanistic models of regulation of the inner membrane channel through which calcium is supplied to mitochondria.
Heme peroxidases are used as powerful biotechnology tools because they catalyze a highly diverse set of reactions. In past work, this team had engineered a monomeric peroxidase, named APEX, derived from dimeric pea plant ascorbate peroxidase. The MIT and UCSD collaborators had already shown that APEX can be expressed in cells without loss of activity so it can be used for intracellular-specific protein imaging by electron microscopy (EM) and spatially resolved proteomic mapping. For EM, APEX can be fused genetically to a protein of interest, and the resulting construct can be expressed inside cells. The cells are then fixed with glutaraldehyde, and a solution of diaminobenzidine (DAB) with hydrogen peroxide is used as the enzyme substrate. APEX catalyzes the local deposition of DAB at the site of the enzyme itself, which then binds electron-dense osmium, providing EM electron-scattering contrast at that location.
For proteomic mapping, APEX is targeted genetically to a cellular organelle or protein complex of interest. Then the host live cells are treated with hydrogen peroxide in the presence of biotin-phenol. APEX catalyzes the oxidation of biotin-phenol to generate a very short-lived radical, which tags proteins proximal to APEX, allowing the proteins closest to the genetically labeled protein to be identified by mass spectrometry.
While APEX has enabled some significant biological discoveries, in practice it suffers from sub-optimal sensitivity: When it is expressed at low levels, its activity becomes undetectable. In some cases, it’s possible to increase its expression, but in other cases overexpression is detrimental. To improve APEX’s sensitivity, the research team employed directed evolution by displaying 106 APEX variants on the surface of yeast cells and screening for the most active colonies. After three rounds of selection, they identified two predominant mutants. As the two exhibited similarly enhanced activity levels, they chose the simpler soybean-derived mutant, which they named APEX2. APEX2 contains the three key mutations in the original APEX probe (K14D, W41F, and E112K), plus an additional A134P modification. First they confirmed that APEX2 was more sensitive than APEX for proteomic mapping. Through microscopy, they observed many cells expressing APEX2 at very low levels that nevertheless had high activity, generating labeling contrast quickly and with high specificity. They also confirmed that APEX2, by comparison with APEX, has a greater ability to mark proximal proteins, a critical measure of its utility for proteomic mapping.
They then progressed to more extensive microscopy studies, confirming that APEX2 is a much more sensitive reporter than the original APEX. DAB staining was performed on fixed cells, which were then imaged by bright-field microscopy. In this mode, cellular regions that appear dark, owing to the absorbance of visible light, reflect the presence and intensity of DAB polymer deposits and, thus, elevated APEX or APEX2 activity. APEX2 fusions consistently reacted more quickly and stained more intensely than their APEX counterparts. Most exciting, though, is that the improved properties of APEX2 allowed the team to investigate and resolve an important biological problem that was previously intractable to study with the original APEX probe.
Mitochondria take up calcium from the cytosol via a channel in the inner membrane of the organelle. The regulation of this channel occurs through a soluble Ca2+ binding protein called MICU1.
Previous conflicting reports localized MICU1 to the intermembrane space (IMS) or the mitochondrial matrix, resulting in very different mechanistic models of regulation. IMS localization implies that MICU1 senses and responds to increases in calcium because the porous outer mitochondrial membrane enables its free exchange between the IMS and cytosol. In contrast, matrix localization suggests that MICU1 senses and responds to matrix calcium alone, as the inner membrane is impermeable to it.
EM imaging showed APEX2 staining exclusively in the IMS and not the matrix ( see Figure above). Because of the high spatial resolution of this approach, the team also observed that MICU1-APEX2 was associated with the inner mitochondrial membrane rather than filling the entire IMS, as expected for a protein that forms a complex with inner membrane channels. This result, coupled with recent proteomic evidence, strongly suggests that MICU1 is localized exclusively to the IMS, confirming the validity of the first of the conflicting reports previously addressing this question.
In summary, three rounds of yeast display evolution produced a single mutant with four key amino acid modifications that greatly enhances the cellular activity and, hence, sensitivity of APEX. Consequently, APEX2 can be used for EM studies and proteomic tagging experiments not previously possible with APEX. As an EM reporter, APEX2 is easier to use than most other tags and can provide 3D staining across large fields of view without special equipment because contrast generation does not require light, as is applied in the photooxidation process. The directness of this approach makes it an attractive option to examine the suborganellar localization and membrane topology of important proteins, including tissues of organisms.
Funding for this work comes from the National Institutes of Health (DP1 OD003961, P41 GM103412, R01GM086197, 5R01GM077465-08), the Howard Hughes Medical Institute, and a Howard Hughes Medical Institute Collaborative Initiative Award.
Citation: Lam, Stephanie S., Jeffrey D. Martell, Kimberli J. Kramer, Thomas J. Deerinck, Mark H. Ellisman, Vamsi K Mootha, and Alice Y. Ting, Directed evolution of APEX2 for electron microscopy and proximity labeling, Nature Methods (Advance Online Publication), November 24, 2014, doi:10.1038/nmeth.3179.
